Abstract
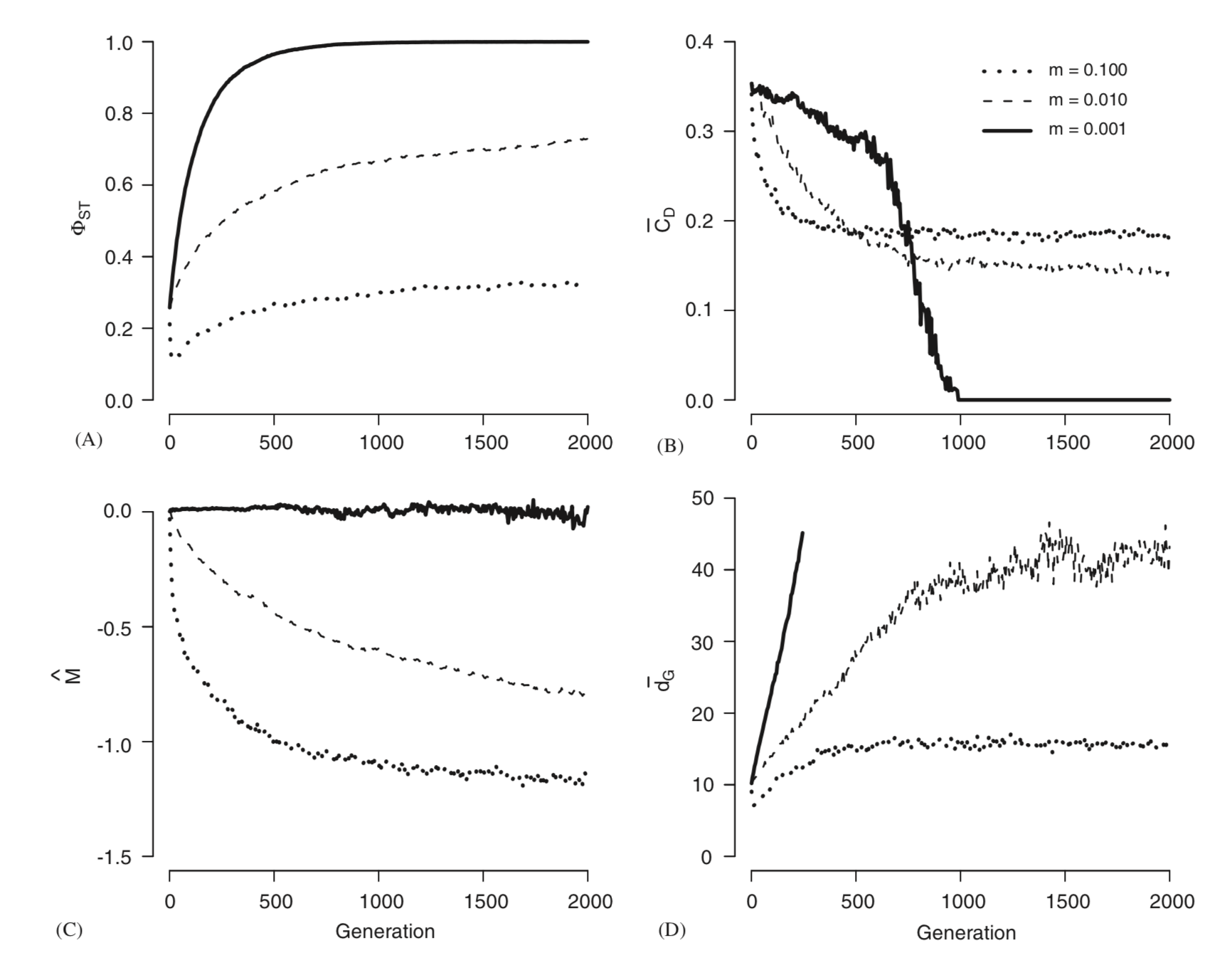
This manuscript explores the simultaneous evolution of population genetic parameters and topological features within a population graph through a series of Monte Carlo simulations. I show that node centrality and graph breadth are significantly correlated to population genetic parameters Phi(ST) and M (p = -0.95; p = -0.98, respectively), which are commonly used in quantifying among population genetic structure and isolation by distance. Next, the topological consequences of migration patterns are examined by contrasting N-island and stepping stone models of gene movement. Finally, I show how variation in migration rate influences the rate of formation of specific topological features with particular emphasis to the phase transition that occurs when populations begin to become fixed due to restricted movement of genes among populations. I close by discussing the utility of this method for the analysis of intraspecific genetic variation. (c) 2006 Elsevier Inc. All rights reserved.

